This is an old revision of the document!
Table of Contents
CrEDIBLE thematic working days, October 8-10, 2014
Due to the increasing on-line availability of various biomedical data sources, the ability to federate heterogeneous and distributed data sources becomes critical to support multi-centric studies and translational research in medicine. The CrEDIBLE project organises 3 thematic working days in October 8-10 in Sophia Antipolis (near to Nice, France) where experts are invited to present their latest work and discuss their approaches. The aim is to gather scientists from all disciplines involved in the set up of distributed and heterogeneous medical data sharing systems (medical data representation, data mediation, data stores federation, data semantics, workflows, … towards biomedical data integration), to provide an overview of this broad and complex area, to assess the state-of-the-art methods and technologies addressing it, and to discuss the open scientific questions it raises.
The methods for biomedical data distribution considered in the context of CrEDIBLE are:
- Modelling: ontology-based extensible data models.
- Federation: the (virtual) fusion of geographically spread data stores which should appear to end users as a unique and coherent data source.
- Mediation: the semantic alignment of heterogeneous data sources, which were often designed independently from each other.
- Querying: the description of distributed data sets, defined through data retrieval queries that apply on the whole federated system.
Two previous issues of this workshops were organised in 2013 and 2012.
Working days organisation
The idea of this workshop it to have groups of ~30 minutes presentations within a given theme/session followed by a time slot for a panel discussion to share the presenters experience on selected scientific questions and challenges. Talks should keep a balance between introducing the field (appropriate for a broad audience of scientists involved in all the areas covered by CrEDIBLE) and technical details (appropriate for expert scientists).
Thematic sessions
Session 1: Biomedical data federation in practice: feedback on existing approaches
Session goal: To report on concrete experience in developing systems gathering or indexing data to be shared and reused in research projects. User requirements, current technology limitations and future expectations.
Scientific questions:
- Data indexing: how to meet the expectations of researchers in terms of precision of the vocabulary
- Data provenance: what do we need actually: detailed models of provenance ? or more “distilled” information ?
- Access control: how to accomodate multiple access policies specified by contributing entities ? is it manageable in practice ? should it be applied to datasets only (e.g. images, signals) or to metadata as well ?
- Data federation: what level of data federation is required? What are the data sources to federate? What are the data models in use?
Session 2: Reasoning and visualizing large data graphs
Context: Once data from heterogeneous and distributed store has been mediated and federated, the question which arises is the exploitation of the large data graph available. This implies, among others, to enable reasoning over it and visualizing it. While the Web of data focuses on large data sets processing, the semantic Web involves costly reasoning processes. There is a trade-off to be found between the amount of data to process and the reasoning capabilities of the system. Meanwhile, information visualization aims at providing methods and tools to access and explore large amounts of data, at conveying knowledge from these data in intuitive ways and therefore understanding them. For the Web of data, we focus on the visualization of large data graphs.
Scientific questions:
- How to visualize and navigate on large data graphs for exploratory search, visual mining and understanding of these data?
- How to apply inference rules on large distributed data graphs?
- How to combine visualization of data and automatic reasoning?
Session 3: Data federation
Context: non-partitioned data stores, no prior knowledge on stores content, common RDF representation, common reference ontology.
Scientific questions:
- Query language. Is SPARQL (v1.1) the most appropriate language? What is the trade-off between expressiveness and performance?
- Performance. What is the performance impact? Gain of parallel execution of queries vs network overhead?
- Especially when deploying over a WAN?
- Scalability. How scalable are the different methods proposed? To what scale have they been tested?
- Reliability. What is the impact of low reliability? Can queries be partially processed in case of communication failures with some data stores? Can end-users be notified on the kind of potentially missing information?
Session 4: Data mediation
Context: heterogeneous data stores, within the same or related domains (e.g. biology, medical images, clinical records…), considering sensitive data.
Scientific questions:
- From a read web of data to a read/write web of data: what solutions are proposed so far? Changes back to the source databases or written apart? What are the future perspectives?
- Query language. SPARQL can be used as query language to access data in heterogeneous databases. Is it the most appropriate query language? What are the query languages applicable to the methods presented.
- How to mediate various data sources (Pros and cons of each approach. Use cases. Are there hybrid approaches?):
- statically (ETL): transform data source periodically (e.g. into RDF stores).
- dynamically: query on-the-fly.
- How to ensure access control in an heterogeneous deployment
- at coarse-grain (each data repository)
- at fine-grain (each entity within a data repository)
- Reference model. Taxonomies or ontologies can be used as reference model. Is this the most appropriate reference model? What are the target models of the methods presented.
Session 5: Biomedical ontologies
Session goal: To discuss ontologies modeling observations and measurements data (designed to facilitate the sharing and reuse of scientific data)
Scientific questions:
- modelling related entities (observed entity, measured quality, measurement results, units of measurements) and relationships
- relation with foundational ontology (ies) such as DOLCE, DOLCE-CORE and BFO ? compatibility ?
- relation to existing ontologies of qualities
- how to model complex observation data such as images ? notion of field ?
- how to model time varying phenomena ?
Agenda
Wednesday, October the 8th, 2014
14:00 - 14:30, Welcome and introduction
- Johan Montagnat (CNRS, FR), Introduction
14:30 - 17:30, Session 1: Biomedical data federation in practice: feedback on existing approaches
- Camille Maumet (University of Warwick, UK): Supporting image-based meta-analysis with NIDM: Standardised reporting of neuroimaging results
- Silvia Olabarriaga, (Amsterdam Medical Center, NL): Challenges for services integration into science gateways: the local story of the AMC
- Pascal Neveu (INRA Montpellier, FR): From Gene To The Bottle
- Frédérique Segond (Viseo, FR): Language and knowledge technologies to properly model textual medical data and support better reasoning
- Panel discussion
Thursday, October the 9th, 2014
9:00 - 11:30, Session 2: Reasoning on Graphs
- Harald Sack (U. Potsdam, DE): The Journey is the Reward - Exploratory Semantic Search based on Linked Data
- Martin Peters (U. Dortmund, DE): Rule-based reasoning using GPUs
- Guy Melançon (U. Bordeaux): Visual Analytics Supporting Rule Based Modeling
- Christian De Sainte Marie (IBM): Business rules engines
- Panel discussion
13:00 - 15:00, Session 3: Data federation
- Ruben Verborgh (Ghent University): Querying data on the Web – client or server?
- Ester Pacitti (U. Montpellier 2 / INRIA, FR): Profile Diversity for Query Processing using Users Recommendations
- Axel-Cyrille Ngonga Ngomo (U. Leipzig): HIBISCUS: hypergraph-based sources selection for federated SPARQL queries
- Panel discussion
15:30 - 17:30, Session 4: Data mediation
- Pascal Molli (U. Nantes, FR): Towards Writable and Scalable Linked Open Data
- Franck Michel (CNRS, FR): xR2RML, an R2RML extension for the translation of non-relational databases into RDF
- Freddy Priyatna (U. Polytechnic Madrid, SP): An Overview of the Research Carried Out at Data Integration Group - OEG
- Panel discussion
Friday, October the 10th, 2014
9:00 - 11:30, Session 5: Biomedical ontologies
- Simon Jupp (EBI, UK): BiomedBridges project
- Heiner Oberkampf (Siemens, DE): Expressing Medical Image Measurements using Open Biological and Biomedical Ontologies
- Alan Ruttenberg (University at Buffalo, School of Dental Medicine, USA): What should the role of biomedical ontology in imaging be?
- Bernard Gibaud (INSERM, FR): ontology modelling needs for image biomarkers description
- Panel discussion
Workshop conclusions
- 11:30, Johan Montagnat (CNRS, FR)
Abstracts
- Camille Maumet (University of Warwick, UK): “Supporting image-based meta-analysis with NIDM: Standardised reporting of neuroimaging results”. Due to the lack of data shared when reporting neuroimaging results, most neuroimaging meta-analyses are based on peak coordinate data. However, the best practice is an image-based meta-analysis that combines full image data of the effect estimates and standard errors derived from each study. The Neuroimaging Data Model (NIDM) is an ongoing effort, supported by the INCF, to provide a domain-specific extension of the W3C PROV-DM. In this talk, I will review our recent progress in extending NIDM to share the statistical results of a neuroimaging study and our interactions with existing software packages (SPM, FSL, AFNI, Neurovault.org).
- Silvia Olabarriaga, (Amsterdam Medical Center, NL): “Challenges for services integration into science gateways: the local story of the AMC”. In this talk I will present and discuss the experience and difficulties faced by our group to build science gateways that integrate data and computation resources for biomedical data analysis and simulation. The various generations of systems developed so far will be briefly presented, highlighting the solutions (or absence thereof) to deal with heterogeneous computation and data services.
- Pascal Neveu (INRA Montpellier, FR): “From Gene To The Bottle”. In agronomy a lot of research questions , such as “How to adapt agriculture to climate change?”, require interdisciplinary work. Agronomy science uses a wide range of disciplines and that is why it poses specific issues in terms of data and knowledge integration. This presentation will give a description of a current work on viticulture and wine.
- Frédérique Segond (Viseo, FR): “Language and knowledge technologies to properly model textual medical data and support better reasoning”. In this talk we will present the work performed in Synodos, a project dedicated to information extraction from Electronic Medical Records. We will first spend some time to present the type of unstructured textual data contained in these records together with a list of difficulties encountered to properly structure them. We will then describe the methodology and the different technologies we use to achieve our goal. This work lies at the intersection of four disciplines: terminology, natural language processing, knowledge representation and Reasoning.
- Pascal Molli (U. Nantes, FR): “Towards Writable and Scalable Linked Open Data”. Current Linked Open Data faces severe issues of scalability, availability and data quality. To solve the data quality issue, data publishers crowdsource to external collaborators, that make partial copies of the dataset and update it. We propose to take advantage of the fragmentation and replication produced by this process to improve availability and scalability by sharing the query load with the collaborators.
- Freddy Priyatna (U. Polytechnic Madrid, SP): “An Overview of the Research Carried Out at Data Integration Group - OEG”. Ontology Based Data Access (OBDA) utilizes a target ontology for accessing the underlying data-sources. In this talk, we highlight the recent research carried out at the Data Integration Group - Ontology Engineering Group, such as query answering (rewriting and translation). Furthermore, we will take an overview of morph-suite, the implementation of our research, that consists of morph-RDB (relational based OBDA), morph-stream (sensor-based OBDA), morph-GFT (web-based storage based OBDA), and morph-LDP (read-write access OBDA combined with our Linked Data Platform implementation). Finally, we will report our experience of using one of the tools to deal with various biomedical queries.
- Franck Michel (CNRS, FR): “xR2RML: an R2RML extension for the translation of non-relational databases into RDF”. The web of data is progressively emerging along with the publication and interlinking of various open data sets in RDF. Its success largely depends on the accessibility of semi-structured documents and structured databases hardly indexed by standard search engines. In particular, NoSQL systems have gained a remarkable success during recent years. Other types of databases have been developed over time, such as XML databases (notably used in edition and digital humanities), object-oriented databases or directory-based databases. Significant efforts have been invested in the definition of methods to translate various kinds of data sources into RDF, such as R2RML, a RDB-to-RDF W3C recommendation. So far however, no common language describes, in a uniform manner, mappings of most common types of databases to RDF. In this presentation we introduce xR2RML, an extension of R2RML that addresses the mapping of a large, extensible, scope of non-relational databases to RDF. In particular we show that xR2RML is designed to flexibly adapt to various data models and query languages.
- Ester Pacitti (U. Montpellier 2 / INRIA, FR): “Profile Diversity for Query Processing using Users Recommendations”. Many scientific fields produce and consume a considerable amount of diverse data (e.g. biology , astronomy, physics) stored in different heterogeneous sites, and produced by different types of users profiles. We investigate two different use cases: a) In the domain of plant phenotyping, there has recently been increasing interests in finding diverse data coming from different research communities. b)In botany, the emergence of citizen sciences has fostered the creation of large and structured communities of nature observers. In this context, there is a need to retrieve diverse plant observations from a diverse spectrum of plant families, genus and species. In this talk I will present some new issues of profile diversity, a novel idea in searching and recommending scientific items (e.g. documents, images, datasets, etc), and how profile diversity can be deployed in different kinds of infrastructures (centralized and distributed).
- Ruben Verborgh (Ghent University): “Querying data on the Web – client or server?”. After 15 years of Semantic Web research, we are still struggling to make things scale. How can we reliably execute complex queries over data on the Web, without having to copy that data to our own database? If we really want “Web” querying, we need a contract between clients and servers with a different balance. This talk invites you to think about new interfaces, starting from a concrete proposal with proven availability, and aims to start a discussion about the involved trade-offs.
- Harald Sack (U. Potsdam, DE), “The Journey is the Reward - Exploratory Semantic Search based on Linked Data”. Today's (web) search engines deliver pinpoint results as long as the user knows how to formulate the appropriate search query. But, if the user is not familiar with the search domain or if the information need of the user requires subsequent queries that build on one another, traditional retrieval technology reaches its limits. Exploratory search exploits the content-based relationship of documents to enable the discovery of relevant results along the guided search path. Moreover, it enables also serendipitious discovery of solutions to a problem that is relevant but not intentionally thought of.
- Martin Peters (U. Dortmund, DE), “Rule-based reasoning using GPUs”. The talk will be about how a rule-based reasoner can be implemented, that uses the massively parallel hardware of modern graphic cards (GPUs). In detail I’m going to show how the RETE algorithm, which is a pattern-matching algorithm that can be used to implement production systems, can be adapted to for a highly parallel execution on the GPU. Based on the introduced concepts the materialization of the complete RDFS closure for 1 billion triples can be performed on a single computing node, reaching a throughput that is comparable to state of the art MapReduce-based approaches.
- Guy Melançon (U. Bordeaux), Visual Analytics Supporting Rule Based Modeling. The talk will go over our efforts in recent years trying to build a visual analytics system supporting rule-based modeling. The PORGY framework we build and distribute supports tasks that were carefully identified as central to modeling with rules. The system also relies on a strategy language steering rule application, and actually being a crucial part of models.
- Heiner Oberkampf (Siemens, DE): “Expressing Medical Image Measurements using Open Biological and Biomedical Ontologies”. Radiological images in include rich and complex information that radiologists must summarize with just a few measurements. It is crucial that these measurements be comparable between patients, to assess abnormalities, and between examinations of the same patient, to track changes in health. We show how radiology measurements can be represented in detail using the Ontology for Biomedical Investigations and other Open Biological and Biomedical Ontologies, then describe how this representation allows us to compare measurements against normal values and across consecutive examinations.
- Alan Ruttenberg (University at Buffalo, School of Dental Medicine, USA): “What should the role of biomedical ontology in imaging be?”. The most common use of biomedical ontologies has been to “annotate”, to attach, to or as part of data, standard identifiers of subject matter or elements of process which can then be used as search keys for browsing and aggregation. However ontology and associated technologies can play a much wider role in general and in imaging in particular. In this presentation I'll contrast annotation with using ontology to structure information and make assertions. I'll then give a sketch of how many types of imaging related information, can be organized in a single framework under the Basic Formal Ontology (BFO) and ontologies built on it, and suggest opportunities for further development.
- Bernard Gibaud (INSERM, FR): “Toward ontologies for imaging biomarkers description”.The presentation will introduce imaging biomarkers, and stress their importance in biomedical research (both in clinical and translational research) as well in key decisions concerning patient management. In this context, the deployment and federation of imaging biobanks should provide the basic infrastructure to support the sharing of biomedical imaging data (including imaging biomarkers), and stimulate research about biomarkers' design, qualification and clinical use. Ontologies and other semantic web technologies could facilitate the sharing of this information. The presentation will review some of the ontology sources currently available or in development, and discuss their relevance (e.g. OBI, RadLex, QIBO, OntoNeuroLog). The most salient open issues will be listed.
Venue
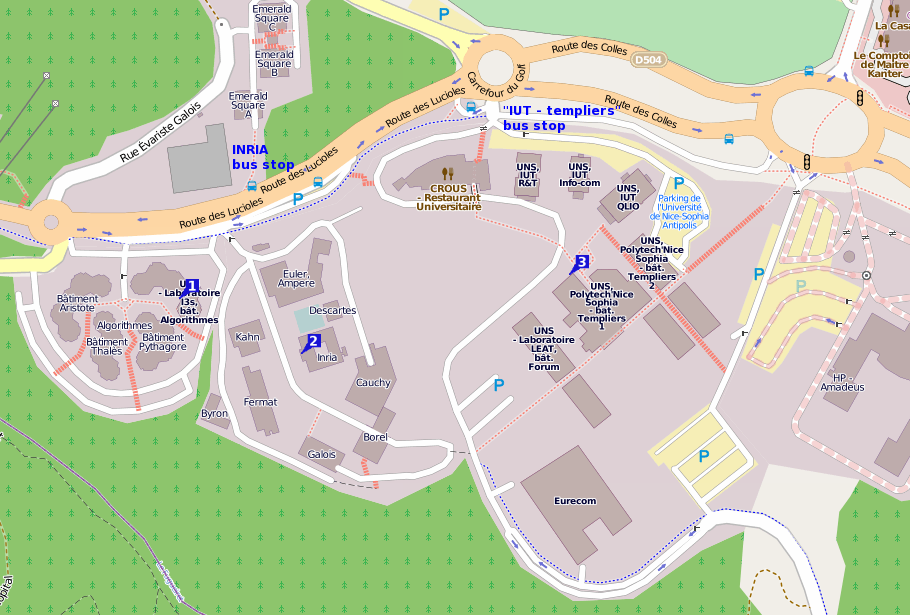
The workshop will be held in the conference room, located at the ground floor of the I3S laboratory, in Sophia Antipolis, France. The I3S laboratory address is: Building “Algorithms B”, 2000 route des Lucioles, BP 121, 06903 Sophia Antipolis Cedex, FRANCE
Where is it ?
- GPS coordinates:
- 43°36'56“ N
- 07°04'01” E
- See number [1] on the map below
How to go there ?
- By plane: "Nice Cote d'Azur" airport (NCE). There are two terminals (T1 and T2) with a free and frequent shuttle bus circling between the terminals.
- Taxi are available from both terminals. A taxi from the airport costs approximately 65 euros.
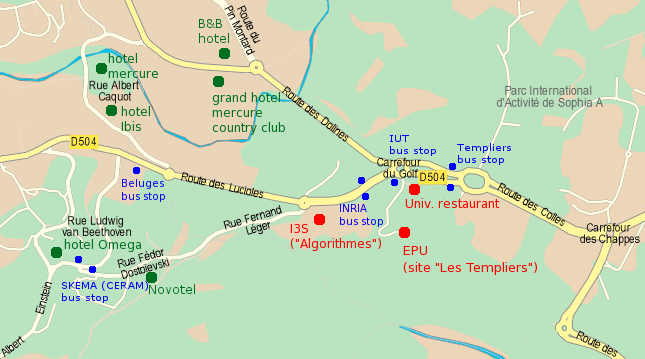
- During week days, from the airport T1 TAM bus number 230 (Sophia Express bus) is direct to Sophia Antipolis through the motorway (stop at the “INRIA” bus stop shown on the map). See the schedule.
- On saturdays, Sundays and Holidays you need to take metropolitan TAM bus number 200 (towards Cannes) and stop at the Antibes train station bridge (“passerelle SNCF” bus stop) before connecting to one of the buses from Antibes to Sophia Antipolis (see below). The buses time tables are sparse on Sundays and Holidays.
- The taxi or bus 230 ride is approximately 20 minutes out of rush hours.
- By train: “Antibes” train station. From the train station there are 3 options:
- Bus Envibus line number 11 is direct from Antibes train station (“SNCF” bus stop) to EPU building (“IUT” bus stop).
- Bus Envibus line number 1 runs from the train station bridge (“passerelle SNCF” bus stop) to Sophia Antipolis (“IUT bus stop”). Be aware that there are 2 buses number 1 line ends: “Lycée Léonard de Vinci” (these ones stops before reaching Sophia Antipolis) and “Gare Routière de Valbonne - Sophia Antipolis” (take one of these).
- Bus Envibus line number 9 runs from bus stop “Vautrin bas” (in the vicinity of the Antibes train station bridge, short walk on your left after crossing the bridge) to the “Belugues” bus stop in Sophia Antipolis (see map). Be aware that there are 2 buses number 9 line ends: “Lycée Léonard de Vinci” (these ones stops before reaching Sophia Antipolis) and “Gare Routière de Valbonne - Sophia Antipolis” (take one of these).
- By car: A8 highway. Exit at “Antibes - Sophia Antipolis” then follow Sophia Antipolis and reach “carrefour des Chappes” round-about on the map below.
- The I3S building is number “[1]” on the map below, which also shows the closest bus stops.
Also see the interactive map.
Nearby hotels
A list of nearby hotels (all within 10 minutes walking distance) is located on this map:
- B&B'hotel:
http://www.hotel-bb.com/hotel_info?hotelId=4532
- Hotel Ibis, 2 stars:
http://www.ibishotel.com/gb/hotel-0711-ibis-antibes-sophia-antipolis/index.shtml
- Hotel Mercure, 3 stars:
http://www.accorhotels.com/gb/hotel-1122-mercure-antibes-sophia-antipolis/index.shtml
- Hotel Omega, 3 stars:
http://www.hotelomega.com/en/index.php
- Grand Hotel Mercure Sophia Country Club, 4 stars:
http://www.accorhotels.com/gb/hotel-1279-grand-hotel-mercure-sophia-country-club/index.shtml
- Novotel, 4 stars:
http://www.novotel.com/gb/hotel-0398-novotel-sophia-antipolis/index.shtml