Table of Contents
Thematic working days, October 15-17, 2012
Due to the increasing on-line availability of various biomedical data sources, the ability to federate heterogeneous and distributed data sources becomes critical to support multi-centric studies and translational research in medicine. The CrEDIBLE project organized 3 thematic working days in October 15-17 2012 in Sophia Antipolis (France) where experts were invited to present their latest work and discuss their approaches. The aim was to gather scientists from all disciplines involved in the set up of distributed and heterogeneous medical image data sharing systems, to provide an overview of this broad and complex area, to assess the state-of-the-art methods and technologies addressing it, and to discuss the open scientific questions it raises.
The methods for biomedical data distribution considered in the context of CrEDIBLE are:
- Federation: the (virtual) fusion of geographically spread data stores which should appear to end users as a unique and coherent data source.
- Mediation: the semantic alignment of heterogeneous data sources, which were often designed independently from each other.
- Querying: the description of distributed data sets, defined through data retrieval queries that apply on the whole federated system.
- Data flow: the use and the enrichment of the federated data stores through the use of data processing pipelines.
Agenda
- Monday, October 15
- 12:00 - Lunch
- 14:00 - J. Montagnat (CNRS) - Welcome, CrEDIBLE project presentation, feedback from the NeuroLOG project
- 14:30 - Session 1: Data integration methods and tools (moderators: Johan Montagnat, Olivier Corby)
- Medical data integration
- 14:30 - C. Daniel (APHP / INSERM, Paris) - Electronic Health Records for Clinical Research
- 15:10 - S. Murphy (Massachusetts General Hospital / Harvard Medical School) - Instrumenting the Health Care Enterprise for Discovery Research
- 16:00 - coffee break
- Data mediation
- 16:20 - O. Corcho (U. Polytechnic Madrid) - Distributed queries, data mediation
- 17:00 - P. Grenon (European Bioinformatics Institute) - Ontology based knowledge management of biomedical models and data
- 17:20 - O. Corby (INRIA, Sophia Antipolis) - KGRAM abstract machine for knowledge data management
- 17:40 - Discussions
- 18:30 - End of first day
- 19:30 - Diner
- Tuesday, October 16
- 8:00 - Session 2: Ontologies, semantic modeling (moderators: Bernard Gibaud, Gilles Kassel)
- 8:00 - C. Masolo (Laboratory for Applied Ontology, Trento) - DOLCE extensions
- 9:00 - G. Gkoutos (U. Cambridge) - From Systems Genetics to Translational Medicine
- 9:40 - Coffee break
- 10:00 - J. Charlet (APHP / INSERM, Paris) - Relations between Ontologies and Knowledge Structure: Two Case Study
- 10:40 - P. Grenon (European Bioinformatics Institute) - Ontology for biomedical models and data
- 11:10 - B. Batrancourt (APHP / INSERM, Paris) - Ontology reuse, from NeuroLOG to CATI
- 12:00 - Lunch
- 13:00 - Session 3: Data representation model and reasoning (moderators: Catherine Faron-Zucker, Olivier Corby)
- 13:00 - M.-A. Aufaure (Ecole Centrale de Paris) - Crunch and Manage Graph Data: the survival kit
- 13:30 - C. Raissi (INRIA, Nancy) - Knowledge Discovery guided by Domain Knowledge in the Big Data Era
- 14:10 - K. Todorov (INRIA, Montpellier) - Bringing Together Heterogeneous Domain Ontologies via the Construction of a Common Fuzzy Knowledge Body
- 14:40 - Coffee break
- 15:00 - R. Choquet (APHP / INSERM Paris) - DebugIT: Ontology-mediated Data Integration for real-time Antibiotics Resistance Surveillance
- 15:30 - S. Ferré (U. Rennes 1) - SEWELIS: Reconciling Expressive Querying and Exploratory Search
- 16:00 - P. Molli (U. Nantes) - Live Linked Data
- 16:30 - Discussions
- 17:00 - End of second day
- Wednesday, October 17
- 8:00 - Session 4: Semantic workflows (moderators: Alban Gaignard, Tristan Glatard)
- 8:00 - F. Lécué (IBM) - Composing and optimizing services in the Semantic Web
- 8:40 - P. Missier (Newcastle University) - Workflows, experimental findings, and their provenance: towards semantically rich linked data and method sharing for collaborative science
- 9:20 - A. Gaignard, N. Cerezo (I3S, Sophia Antipolis) - Semantic workflows: design and provenance
- 10:00 - Coffee break
- 10:30 - Conclusions
- 12:00 - End of workshop, Lunch
Location
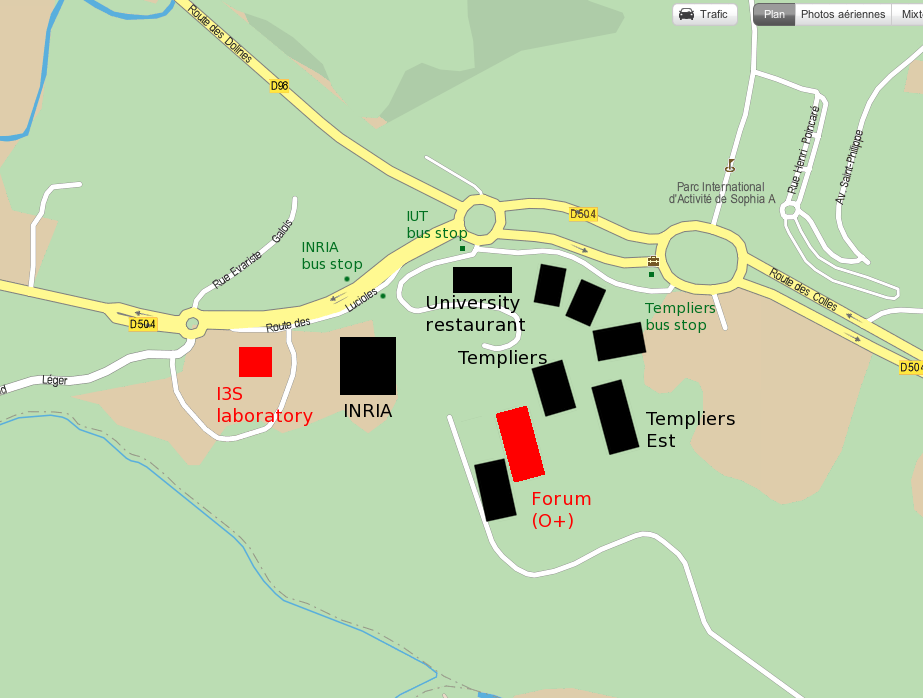
On Monday afternoon, Tuesday afternoon and Wednesday morning, the workshop will be held in the conference room of the I3S laboratory in Sophia Antipolis (France). See the following map to locate the laboratory. The conference room is located at the ground floor of I3S (room number 007).
On Tuesday morning, the workshop will be held in room 101-001 on the first floor of the “Templiers Ouest” building. Please refer to the map below for identifying the building.
Content
Web semantic technologies play a critical role to represent, interprete and query data, to finally achieve data mediation. The themes of knowledge modeling through ontologies and the reuse of existing ontologies will be addressed more specifically. The challenges related to the integration and the federation of heterogeneous data bases, including the data representation models and their impact on the system performance, will be studied. Downstream, the study will also consider the exploitation and the production of new knowledge in the context of data processing workflows. Some feedback on existing tools and their capabilities / limitations is also expected.
Data integration
The data sources to be integrated are related but yet heterogeneous, using different semantic references (vocabularies…), different representations (files, relational / triple / XML databases…) and even different data models (relational, knowledge graphs…). Data integration will also be constrained by medical application constraints, in particular the set up of multi-centric studies, the support of translational research and medical applications. Data security and fine-grained access control is another important related problem. The ability to simultaneously process different data representation model makes data security a particularly challenging problem.
Ontologies
The ontology defines conceptual primitives which represent data semantics (images, test and questionnaire results) by integrating their production context (study, examination, subject, medical practitioner, data acquisition protocol, processing, acquisition device, parameterization, scientific publications). Such an ontology spans over different domains (different entity classes) and includes hundreds of concepts. It is structured through modules with different abstraction levels, to leverage generic primitives that can formalize several domains for practical reason related to ontology maintenance.
The ontology design involves: reusing (completely or partially) existing ontological modules (at different abstraction levels); designing new modules (in particular to represent knowledge related to particular medical domains); managing modules life cycle; documenting to ease reusability. There are different means of exploitation: ontological alignment to federate data that rely on different semantics (addressing problems related to the level of details or even discrepancies in the entities considered); data processing assistance (checking the compatibility of data with processing tools, producing data provenance information); query-based and/or visualization-based data access. Each usage scenario might involve adapting the ontology representation to the tool manipulated (inference engine, visualizer) and its language.
Data representation models and reasoning
Usually, medical data are stored in relational databases which allow for a fast access to data, while metadata are formalized through graph-based knowledge representation models, designed for the semantic Web, which enable reasoning capabilities through inferences based on ontologies used to model this knowledge. Main challenges are the mixed use of different representation and the scalability of data storage and reasoners. The scalability problem is well known in the Web of data community. Promising approaches lie on the use of graph-oriented databases, the adaptation of inferences performed to the size of manipulated data stores, and on querying and reasoning techniques adapted to distributed stores.
Semantic workflows
The acquisition and the representation of knowledge related to the manipulated data is tightly linked to the data processing and transformation tools applied. Knowledge acquired on data may be used to validate or filter the processing tools applied on this data. Conversely, knowledge acquired on processing tools can be used to infer new knowledge on data, in particular the data produced through this processing. Knowledge exploitation can happen at different levels of the scientific processing pipelines life cycle: at design time through editing assistance (static validation, assisted composition) and at run time (dynamic validation, new knowledge creation).
Knowledge on both data and processing tools is also often used to describe data provenance information. Provenance is then described as semantic annotations tracing the execution path. Provenance is tightly related to the nature of data processed. It facilitates the reuse and the interconnection between data from different sources. It can make use of several domain-ontologies and facilitate interoperability between different data processing engines.
List of scientific questions raised
The list of scientific questions of interest for the workshop, classified by session, is shown below.
Questions related to data integration
- How to integrate data structured using different vocabularies?
- What are requirements and advisable solutions to implement:
- multi-centric studies?
- translational research?
- medical applications?…
- How to generate semantic annotation associated to data?
- annotations may be natives, statically generated, or
- dynamically produced by the mediation system from data available in heterogeneous databases or semantic repositories
- define entities targeted by annotations (known as 'data and model resources' in ref RICORDO) such as image series, signals set, clinical research study, medical observation, score obtained when performing a test
- connection of annotation with physical storage of data (files)
- Data access control
- Solutions are usually available to control access to file. Fine-grained access control to relational databases or knowledge graphs is a trickier problem.
Questions related to ontologies
1) Foundational ontologies (DOLCE)
- Ontology adaptability: reusability requires modifications?
- Enrichment with new concepts and foundational relations (e.g. capacity, function, artifact, collection, organization)
- Alignement of foundational ontologies (e.g. DOLCE and BFO): are there techniques to align foundational ontologies and facilitate reusing ontologies grounded on such foundational ontologies?
2) Application ontology (overall project ontology)
- Ontology modular structure: what structure may facilitate reuse of existing ontologies (designed and maintained by other organizations), and favor the reuse of ontology modules in other projects?
- Reuse of existing ontologies: what part can be reused (depending on the project need) and what level of reuse (what needs to be modified to take into account project needs and the overall ontology coherency)? For instance, FMA (Foundational Model of Anatomy) describes normal anatomy. how can it be reused to represent pathological structures? Is the Hoendorf 2007 solution acceptable?
- Creation of new modules, usually to cover the medical areas considered: how to organize such a multidisciplinary work?
- Expressiveness of representation languages: how to properly manipulate relations between Universals (e.g. the head is part of a person; the left hemisphere and the right hemisphere compose part of the brain) and relations between Universals and Individuals (e.g. the hydrogen concentration of a solution, the processing class performed by a software tool), especially at the operations level, to implement inferences (see OWL dialects)?
- Ontology maintenance: versioning, extensions, documenting, public wide spreading?
3) Ontologies alignment
- Choosing an alignment method / an alignment language and associated expressiveness
- Properties of an ontology transformations based on those of its alignment
- Ability to dynamically align concepts at query evaluation time
Questions related to data representation models and reasoning
- Publication and interconnection of linked semantic data sources (Web of data) from raw data sources with heterogeneous structures (e.g. project DataLift)
- Reasoning in graph-based models: large graphs matching
- Incremental approches, approximations
- Data distribution / reasoning distribution
- Visualization
- Storage through NoSQL databases
- Triplestores, e.g. Jena
- Graph-oriented databases, e.g. Neo4j
- Column-oriented databases, e.g. HBase/Hadoop
- Impact on performance
- Impact of the data representation model on security
Questions related to semantic workflows
- Exploiting knowledge on scientific workflows
- At design time: edition assistance, checking, automated composition
- At run time: dynamic validation, new knowledge production
- Provenance
- Producing semantic execution traces, Linked Data (publication, exchange)
- Managing amount and granularity of provenance information
- Domain-agnostic versus domain-specific provenance
- Provenance information and foundational ontologies (Provenir → BFO, OPM → /, Prov-O → ?)
- Extending provenance information to knowledge databases management platform (not restricted to workflow execution, e.g. upon data importation/exportation)
- Interoperability
- Two-ways mapping between standard provenance models (OPM → Prov-DM) and workflow languages for interoperability (e.g. Missier2010).